
Milestones In Marine Microbiology
Marine microbes are abundant and diverse. But their tiny size means they are not easy to study. First observed in 1675, marine microbes were not cultured in the lab for over 100 years allowing for further observation. Molecular advances and large DNA libraries allowed for many advances since the 1970s and there is still much to learn!
Thanks to David Karl and Jody Deming for review of this timeline.
This work was supported by the National Academies Keck Futures Initiative of the National Academy of Sciences under award number NAKFI DBS17. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Academies Keck Futures Initiative or the National Academy of Sciences.
First observations of aquatic microbes
The Dutch lensmaker Antonie van Leeuwenhoek devises a simple, yet powerful single lens microscope that allows him to observe microbes for the first time. He calls the small creatures "wee animalcules."


First attempts at cultivating microbes from the ocean
Adolph-Adrien Certes, a student of Louis Pasteur (the French microbiologist known for his breakthrough work on vaccinations and, of course, pasteurization), reports on his deep-sea microbial cultivation experiments from the French Travailleur and Talisman expeditions.

Publishing of the earliest known book specifically dedicated to marine microbes
The German microbiologist Bernhard Fischer publishes his treatise Die Bakterien des Meeres (Bacteria of the Sea).

The cover page of Die Bakterien des Meeres. Credit: Marine Microbiology: A Monograph on Hydrobacteriology
van Niel begins his summer course in microbiology at Stanford University's Hopkins Marine Station
Running until 1962, Cornelis Bernardus van Niel influences generations of microbiologists with his Hopkins Marine Station summer microbiology course, bringing the Dutch techniques of enrichment culture (a method that allows for the growth of a specific microorganism) to the U.S. and highlighting the tremendous metabolic diversity of microbes in the environment.

ZoBell publishes seminal marine microbe text
Claude ZoBell, often known as the "father of marine microbiology," publishes the foundational text Marine Microbiology: A Monograph on Hydrobacteriology.

Radiotracers first applied to study the activity of marine microbes
The discovery, by Martin Kamen and Sam Ruben, of a radioactive isotope of carbon (14C) allows oceanographer Einer Steeman-Nielsen to follow the uptake of carbon into microbial cells. Scientists begin to quantify the activity of microbes at the base of the food web without having to first culture them in the lab.

Establishment of the Microbial Ecology course at the Marine Biological Laboratory
Holger Jannasch begins the summer Microbial Ecology course (now the Microbial Diversity course) at the Marine Biological Laboratory in Woods Hole, Massachusetts.

The Marine Biological Laboratory in Woods Hole, MA as it appeared sometime between 1930 and 1945. Photo Courtesy of Boston Public Library.
Importance of marine microbes in marine food webs and dissolved material cycling first recognized
Lawrence Pomeroy publishes his work on the role of microbes in the ocean's food web, later dubbed "The Microbial Loop."


Analysis of small subunit ribosomal RNA gene (rRNA) sequences are used to describe the relatedness of organisms leading to the first description of the archaea
Carl Woese and George Fox first propose using rRNA gene sequences as molecular markers for determining the relatedness and organization of life, including marine microorganisms. Ribosomal RNA gene cloning and sequencing from the environment would later be applied to uncultivated microbes in hydrothermal vent symbionts (in 1984) and lead to the discovery of some of the most abundant microbes in the ocean.
Cultivation of the first barophilic (or pressure-adapted) microorganisms from the deep sea

Nuclepore filters and fluorescent staining are used to count marine microbes
Discovery of hydrothermal vents in the deep sea, along with bacteria that thrive only in these high-pressure, high-temperature environments
An expedition to the Galapagos Rift—where tectonic plates slowly spread apart—provides the first visual confirmation of hydrothermal vents, pouring out heated water full of minerals, and a surprise discovery that dense ecosystems of previously unknown organisms are supported by the vent fluid chemistry. Many of these animals depend on symbiotic relationships with bacteria that harvest energy from chemicals like hydrogen sulfide released from the vents. In a follow-on expedition to the East Pacific Rise—the fastest seafloor spreading zone—black smokers are discovered, releasing fluids at an extraordinary temperature of 250°C/482°F (kept liquid by the high pressure of the deep sea). This leads to the discovery of deep-sea hyperthermophiles—organisms that thrive in extremely hot environments—living in a subseafloor biosphere.


Discovery of the abundant marine cyanobacterium Synechococcus
Tiny fluorescent cells observed by John Waterbury on an expedition to the Arabian Sea introduces researchers to Synechococcus. Since then, they have been found in large quantities in almost all ocean water, serving as an important base of the food web for fish and larger mammals.
Discovery of the abundant marine cyanobacterium Prochlorococcus
Sallie (Penny) Chisholm discovers this tiny but super abundant photosynthesizer— estimated to be more abundant than any other on the planet, and responsible for producing 20 percent of the oxygen released to the atmosphere every year.
Establishment of the Hawai'i Ocean Time-series and the Bermuda Atlantic Time-series Study
Two long-term ocean monitoring programs are established—the Hawai'i Ocean Time-series at Station ALOHA by David Karl and Roger Lukas and the Bermuda Atlantic Time-series Study by Anthony Knap. Both programs capture regular physical and biological measurements in order to better understand nutrient cycling, where microbes play a huge role. The information is also important for tracking the impact of climate change and ocean acidification on marine microbes.

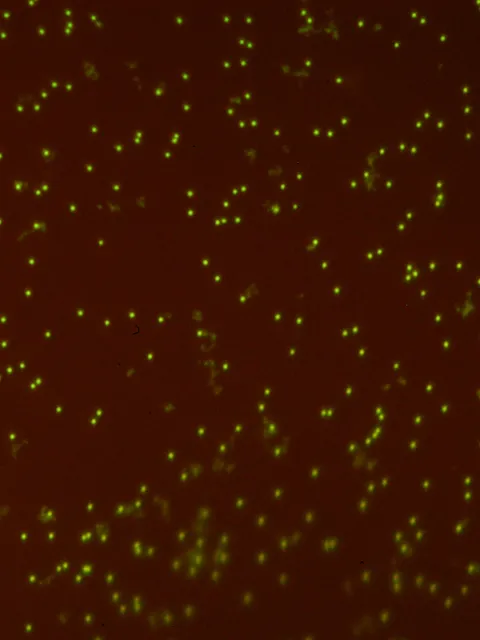
Abundance of viruses in aquatic environments first described
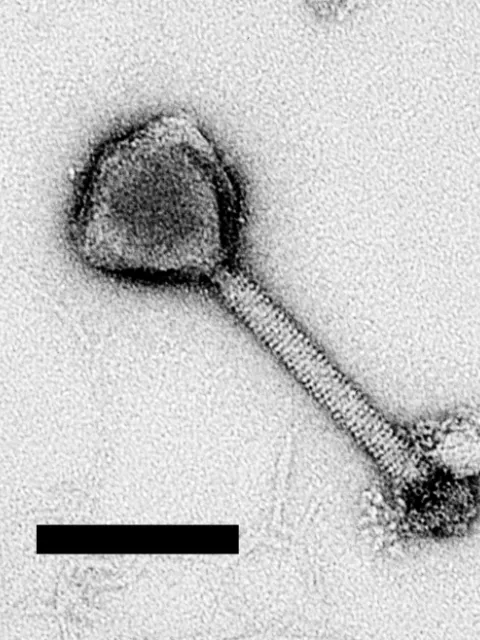
Molecular cloning and DNA sequencing is first applied to planktonic marine microbes and leads to discovery of SAR11, one of the most abundant bacteria in the ocean
Microbes in the SAR11 clade (SAR stands for the Sargasso Sea where they were first discovered by Stephen Giovannoni and colleagues) are the most abundant organic carbon-eating bacteria in the ocean and can make up to 25 percent or more of microbial cells at any given time. DNA sequencing and the ability to clone molecular structures was necessary for the discovery of SAR11 as no cultures of the organism existed at that time.
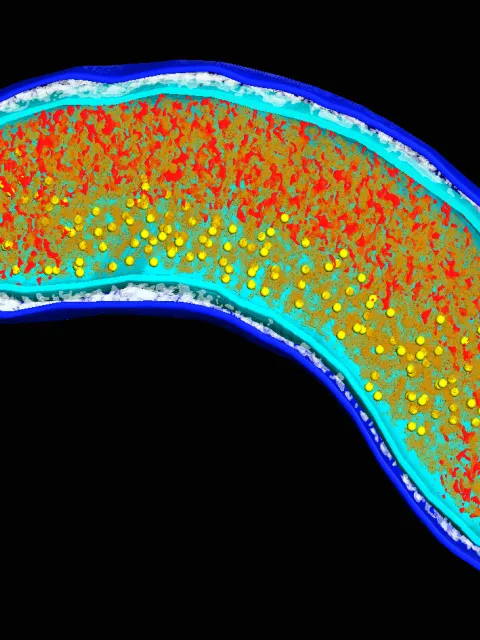

Mesophilic archaea discovered in the marine water column
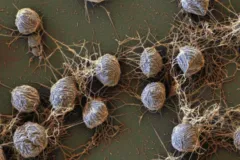

Large-insert DNA libraries are first applied to study metabolic function of uncultivated microbes in the ocean, leading to the discovery of a new type of energy harvest (or phototrophy) in the sea
Discovering microbes by trying to culture them is not easy. Instead, scientists use DNA sequencing to identify just the presence of the microbes' genes without first having to culture them. An early method for doing this involves cloning large pieces of environmental DNA into easily grown E. coli bacteria. Eventually, these techniques lead to the discovery of proteorhodopsin, a protein related to one found in the human eye, which harvests light energy and had not previously been found in bacteria.
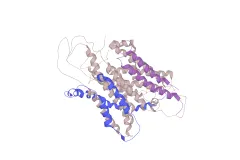
Publication of first complete genomes of marine microbes
The full genomes of several marine microbes including Prochlorococcus, Synechococcus, Pirellula, Silicibacter pomeroyii, the cold-adapted bacterium Colwellia psychrerythraea, and the diatom Thalassiosira pseudonana are published ushering in the genomic era of marine microbiology.
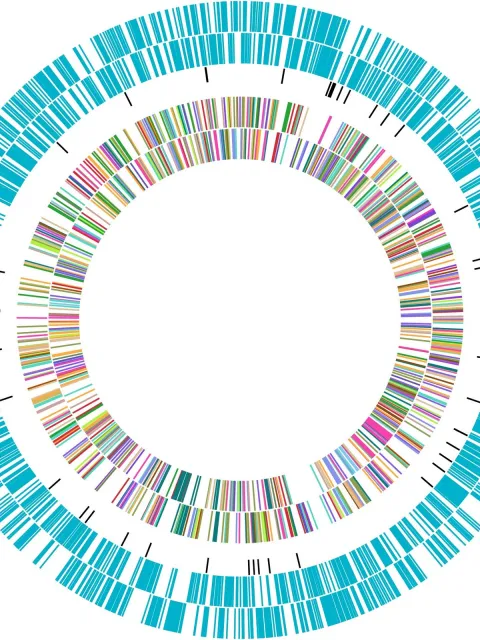
A fuller picture of the microbes in a marine environment are seen using metagenomics
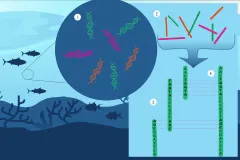
Global Ocean Sampling, Tara Oceans, and Malaspina Expeditions
As it becomes more apparent that marine microbes play critical roles in the marine environment, extensive expeditions that cross multiple ocean basins prioritize studying the microbial world. In 2003, the Global Ocean Sampling Expedition samples 41 locations spanning a distance of 8,000 km (almost 5,000 miles) from the Atlantic to the Pacific, collecting an unprecedented amount of information on marine microbial diversity and abundance. Then in 2010, the Malaspina Expedition circumnavigates the globe, researching the metabolic diversity of microbes in the deep-sea. The Tara Oceans Expeditions spans 2009 to 2013. Crossing the globe, they make monumental strides in understanding the diversity of microbial life in the ocean.

Discovery of Lokiarchaeota; believed to be most recent common ancestor of eukaryotes
Lokiarchaeota found in a deep-sea hydrothermal vent show an evolutionary link between archaea and the more complex eukaryotes. After examining the archaeal DNA, researchers discover that these microbes share about 100 genes for complex cellular functions with eukaryotes, suggesting they are the closest living prokaryotic relatives of eukaryotes.



Connecting the dots


